Dr. Sun Chang, a faculty member of the School of Electronic and Communication Engineering and Artificial Intelligence at Tianjin Normal University (TNU), has published a paper as the first author titled "DCGCN: Dual-channel Graph Convolutional Network-based Drug-target Interactions Prediction Method with 3D Molecular Structure" in the Journal of Chemical Information and Modeling.
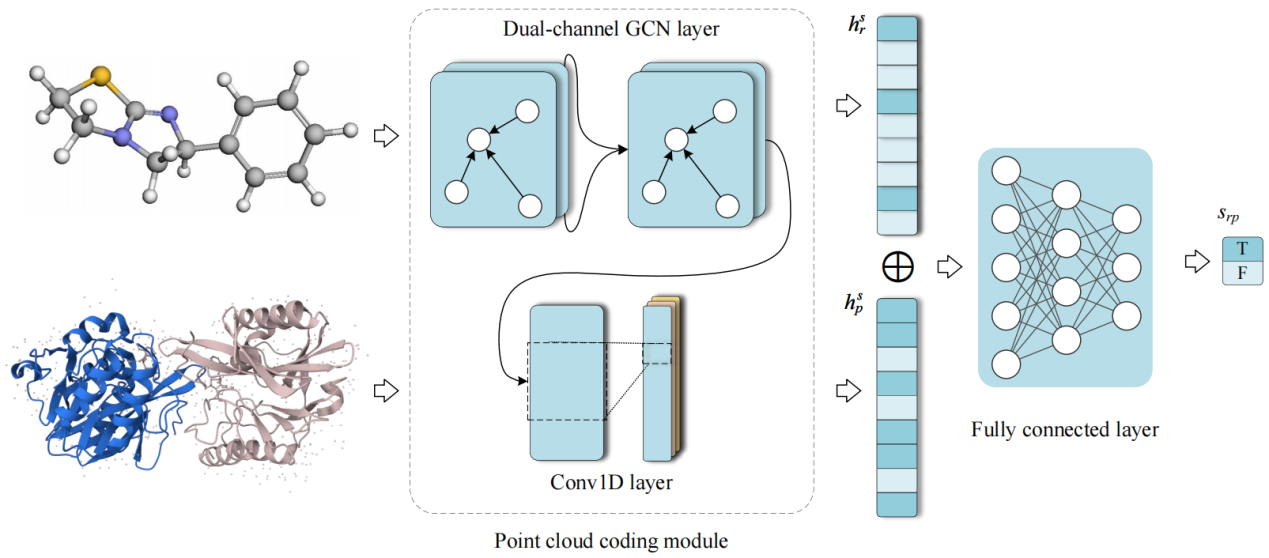
Investigating drug-target interactions (DTIs) is crucial for drug discovery. Most current methods for predicting DTIs rely solely on linear molecular representations, such as SMILES strings or amino acid sequences. However, these linear features fail to adequately capture molecular substructures or the relative positions of atoms. Although some approaches have begun to utilize two-dimensional molecular structures (e.g., bond-line formulas or atomic graphs), they still cannot fully represent a molecule's chemical architecture. To address this, the paper proposes DCGCN, a novel DTI prediction method based on three-dimensional (3D) molecular structure. DCGCN decomposes a molecule's 3D point cloud data into three components: atomic sequence, atomic connectivity, and distance graph. This approach enables the model to better learn the molecular composition of drugs and targets, as well as the connectivity and spatial distances between atoms or amino acid residues.
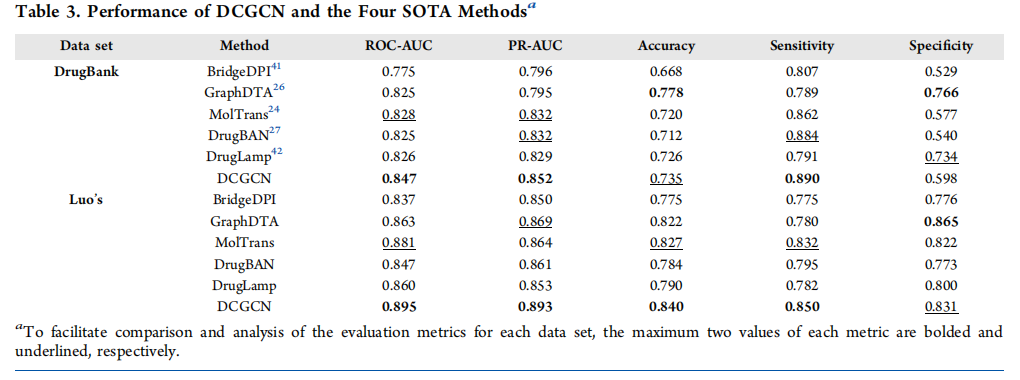
Experimental results on two public datasets demonstrate that DCGCN outperforms several state-of-the-art DTI prediction methods in overall performance. This superiority can be attributed to the fact that 3D molecular structure provides a more comprehensive representation of molecular properties than linear representations. Results from ablation studies confirm that 3D structural information offers more valuable insights. Furthermore, DCGCN's strong performance in cold-start settings validates its effectiveness in identifying novel uses for existing drugs and targets.
A case study shows that DCGCN can leverage known DTIs to identify potential new interactions by comparing the chemical structures and interaction patterns of candidate drugs with those of known ligands. Therefore, DCGCN serves as a practical tool to help biologists screen reliable drug-target candidates, improve the efficiency of biological validation experiments, and accelerate drug development.
Dr. Sun Chang is the first author of the paper, and TNU is the primary affiliation. This research was completed in collaboration with researchers from TNU and Tianjin Fifth Central Hospital. The work was supported by the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 62401392).
Article Links: https://pubs.acs.org/doi/full/10.1021/acs.jcim.5c01012
By He Jierui